Sergey Suchkov1,2,3,4,5,6,7, Afaf El-Ansarya8,9, Amina Al-Haidan8, Veronika Polyakova7, Laila Al-Ayadhi9,10, Lydia Kadyrova4 and Wail M. Hassane11
The Russian Academy of Natural Sciences (RANS), Moscow, Russia
Department of Clinical Allergology & Immunology of the Russian University of Medicine, Moscow, Russia
New York Academy of Sciences, USA
EPMA, Brussels, EU
PMC, Washington, DC, USA
SPM, Tokyo, Japan
The University of World Politics & Law, Moscow, Russia
Autism Center, Lotus Holistic Alternative Medical Center, P.O. Box: 110281, Abu Dhabi, UAE……
Central Research Laboratory, Female Campus, King Saud University, Riyadh, Saudi Arabia
Autism Research and Treatment Center, Riyadh, Saudi Arabia
Department of Biomedical Sciences, University of Missouri Kansas City, School of Medicine
Received: 30 November 2023; Accepted: 10 December 2023; Published: 28 December 2023
Citation: Sergey Suchkov, Afaf El-Ansarya, Amina Al-Haidan, et al. “The Promise of Personalized & Precision Genomics in Autism.” Immunol Res Immunother (2023): 102. DOI: 10.59462/IRIT.1.1.102
Copyright: © 2023 Suchkov S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Despite recent advances in understanding the etiology of autism spectrum disorder (ASD), an approved and efficacious treatment strategy has yet to be identified. The latter demonstrates that the canonical ASD profiled care is becoming much less beneficial and productive, yet it remains ineffective in preventing or effectively treating the above-mentioned disorder. Meanwhile, the link that might exert reliable control over morbidity, mortality and disabling rates as well as significantly optimize the efficacy of ASD treatment for those who had fallen ill and for persons-atrisk is Personalized & Precision Medicine (PPM). Recent advances in ASD genetics pave the way for implementation of PPM in clinical management of autism. Thus, identification of gene modifiers and epigenetic, transcriptional, and translational regulators of implicated genes is important for a more complete understanding of the genetic risk and the precise roles of specific variants.
The inherent complexity and heterogeneity of ASD, often further complicated by comorbid sleep disturbances, epilepsy, or attention deficit hyperactivity disorder (ADHD), has been a prominent impediment to achieving a well-defined treatment algorithm. However, the protocols of symptomatic treatment are not considered in this article, since they have been known for a long time and do not reveal the ideology of personalized targeted therapy. In this review, we have accumulated the latest data from the main areas of pathogenetic target discovery using PPM-based tools and omics technologies, in particular genomics, which hold great promise for the development of personalized preventive diagnostic and therapeutic protocols.
Autism spectrum disorders • Biomarkers • Personalized precision medicine • GABA • Glutamate • Mitochondrial dysfunction • Gut microbiota.
Genetic factors play a significant role in its development of ASDs, and numerous genes contribute to the risk of developing autism, and are thus considered to be crucial for unraveling its causes and developing effective interventions [1]. In reality, autism is considered a complex disorder influenced by the interaction of multiple genes and environmental factors. And thus, multiple genes are likely to converge on shared pathways, resulting in a similar ASD phenotype. The latter, in turn, stem from a disruption of the sophisticated regulatory processes involving differential activity of multiple genes over the course of development. Early-childhood ASD biomarkers are essential both as a primary diagnostic tool and as a means of categorizing patients according to the pathophysiology and etiology of their symptoms.
The core areas of ASD biomarkers research that have emerged during the past decade include biomarkers of GABAergic/glutamatergic imbalance, mitochondrial dysfunction, hyperserotonemia, and impaired gut microbiota. Although many biomarkers have demonstrated impressive successes in distinguishing ASD patients from typically developing children in small studies, such biomarkers have not yet been incorporated into an implementable diagnostic algorithm. Given the heterogeneity of ASD, an ideal diagnostic algorithm should be not only being able to identify ASD patients but should also classify them into etiologic categories with high precision, thus enabling a personalized and precision care approach [2].
Meanwhile, the number of implicated genes far exceeds the number of clinically defined phenotypes, which would show promise for the pre-early identification of ASD. Further deciphering the specific causative role of various genetic and phenotypic perturbations may enhance mechanistic understanding of the disease syndrome and further improve potential treatment options. And thus, the biomarker-driven targeted therapeutic interventions are showing great promise among ASD children, whilst significantly improving care for ASD patients [3]. Through family studies, GWAS, NGS, CNVs analysis, and investigating known autism-associated genes, practitioners and bio designers have gained valuable insights into the genetic underpinnings of ASD. And advancements in technology and PPM-driven approaches hold great promise for developing personalized interventions and improving treatment outcomes for individuals with autism. By integrating genetic information with clinical data, researchers can tailor interventions based on an individual’s unique genetic makeup and specific needs [4].
The power of PPM-driven approach derives from the integrative analysis of multidimensional datasets, including diverse modalities. And thus, PPM as being the Grand Challenge to forecast, to predict and to prevent, is rooted in a big and a new science generated by the achievements of systems biology and translational medicine, whilst integrating OMICS- technologies and Bioinformaticsdriven services, developing an upgraded strategy based on specific biomarkers, including diagnostic, predictive, prognostic, safety, on-treatment and rehabilitationrelated ones. So, a combination of genomic, proteomic, metabolomic and interactomic biomarkers are becoming of great significance and being motivated to be translated into the daily ASD practice to predict risks of the chronification and thus of disabling. This review summarizes the current advances in ASD biomarkers and discusses the potential implementation of genomics, genomics-related technologies and genomics biomarkers in PPM-affiliated areas in the context of ASDs, whilst stressing the impact of PPM as the Science and ART and illustrating application of the different tools of the next step generation at the population, community and individualized levels.
PPM-driven approach in ASD care
According to the National institute of Health (NIH), the definition of Personalized Precision Medicine (PPM) (Figures 1a and 1b) is a new insight of health care in which interventions is informed by each patient’s characteristics, including the genotype, phenotype, and lifestyle [5] as well to enable risk assessment, diagnosis, prevention, and therapy specifically tailored to the unique characteristics of the individual, thus enhancing quality of life and public health. This concept has been adopted in all areas of medicine and healthcare services, acknowledging individual differences. Moreover, the promise of PPM has undergone a global transformation, prompting a concept and philosophy and clinical practice of biomarkers to move ahead from an esoteric realm of pathologists into the lexicon of most physicians, neurology-related practitioners, patients, and even persons-at-risk [6].
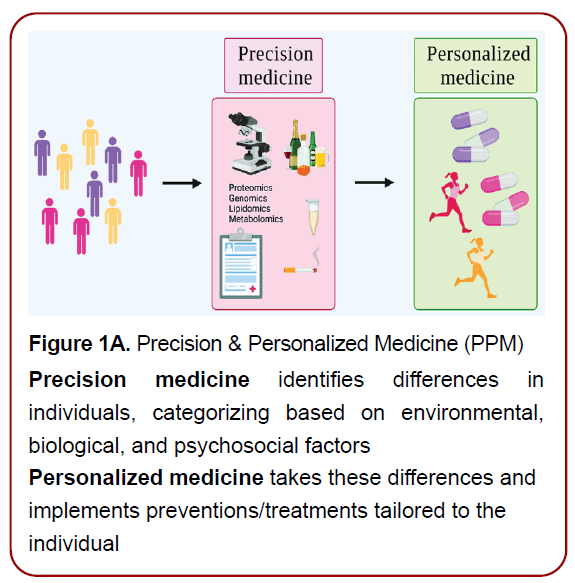
Figure 1A. Precision & Personalized Medicine (PPM)
Precision medicine identifies differences in
individuals, categorizing based on environmental,
biological, and psychosocial factors
Personalized medicine takes these differences and
implements preventions/treatments tailored to the
individual
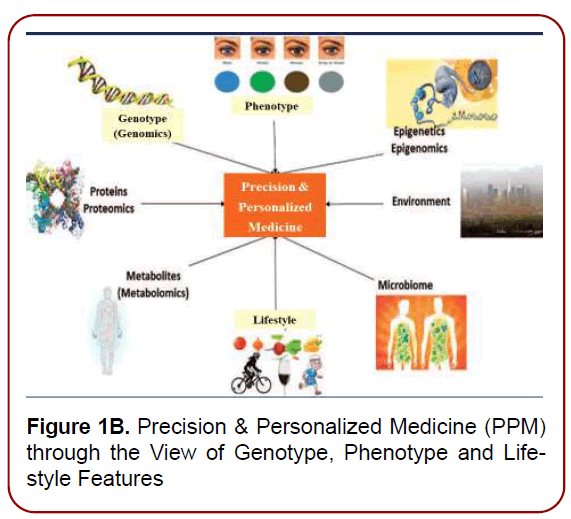
Figure 1B. Precision & Personalized Medicine (PPM) through the View of Genotype, Phenotype and Lifestyle Features
PPM as being the Grand Challenge to forecast, to predict and to prevent is rooted in a big and a new science generated by the achievements(Figure 2a and 2b ) of whilst integrating platforms of Fundamental Sciences and OMICs Technologies (Figure 3), As well as Bioinformatics, Artificial Intelligence (AI), Machine Learning (ML) & Blockchain algorithms (Figure 4) Which are being integrated as into the daily medical practice to secure Clinical, Subclinical & Predictive manipulations of the next step generation.
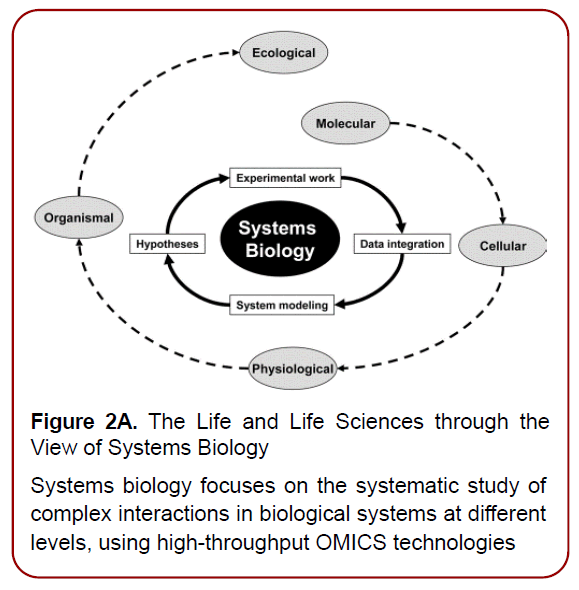
Figure 2A. The Life and Life Sciences through the
View of Systems Biology
Systems biology focuses on the systematic study of
complex interactions in biological systems at different
levels, using high-throughput OMICS technologies
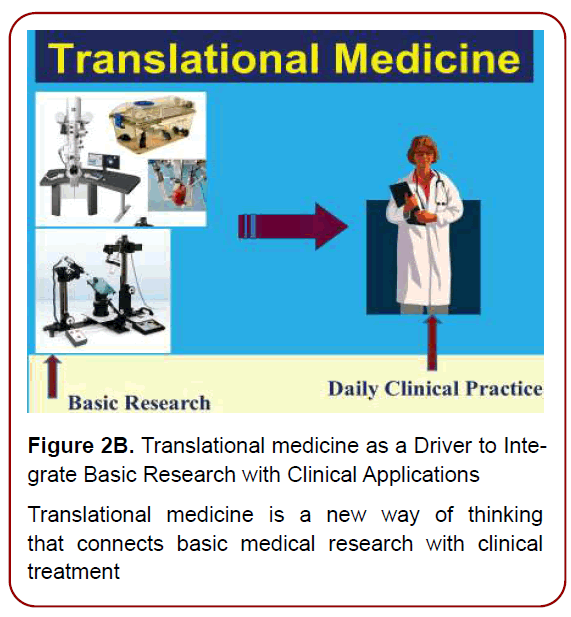
Figure 2B. Translational medicine as a Driver to Integrate
Basic Research with Clinical Applications
Translational medicine is a new way of thinking
that connects basic medical research with clinical
treatment
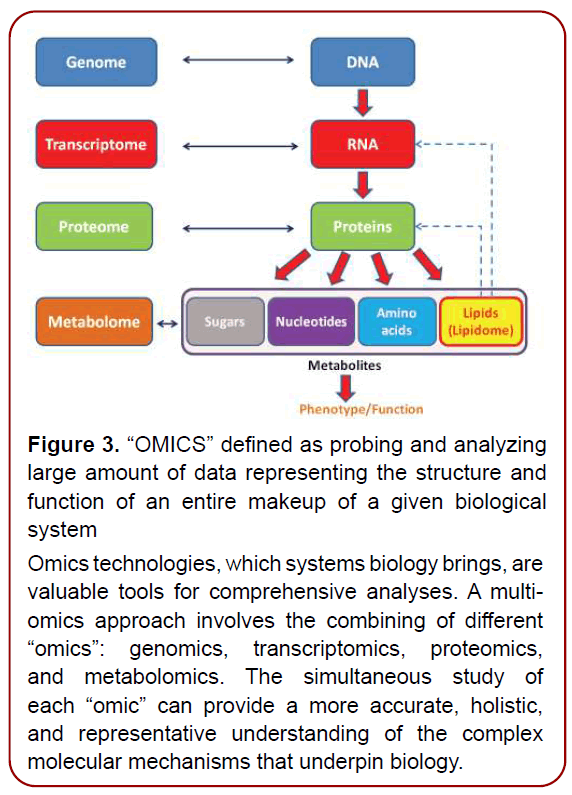
Figure 3. “OMICS” defined as probing and analyzing
large amount of data representing the structure and
function of an entire makeup of a given biological
system
Omics technologies, which systems biology brings, are
valuable tools for comprehensive analyses. A multiomics
approach involves the combining of different
“omics”: genomics, transcriptomics, proteomics,
and metabolomics. The simultaneous study of
each “omic” can provide a more accurate, holistic,
and representative understanding of the complex
molecular mechanisms that underpin biology.
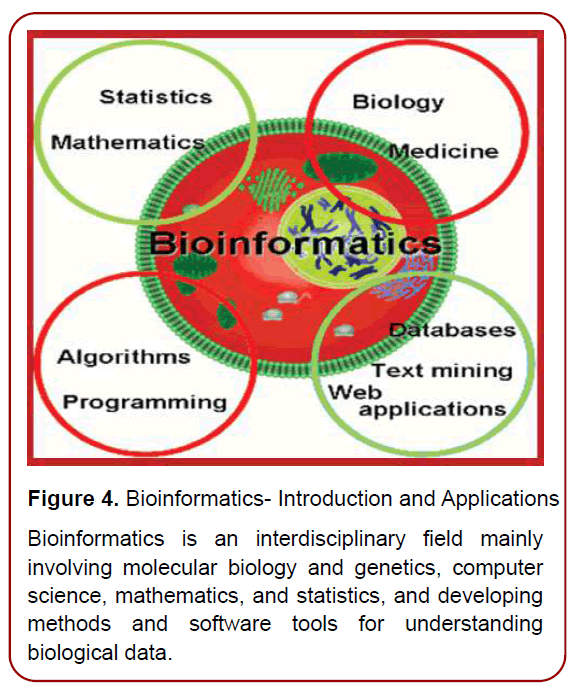
Figure 4. Bioinformatics- Introduction and Applications Bioinformatics is an interdisciplinary field mainly involving molecular biology and genetics, computer science, mathematics, and statistics, and developing methods and software tools for understanding biological data
Moreover, PPM is a goal of healthcare, in which diagnostic and treatment decisions are informed by each person’s unique Clinical, Basics (OMICS-) and Environmental (Exposomics-related) information (Figures 5a and 5b) The success of PPM and its implementation into the practice depend on having accurate diagnostic tests that identify patients who can benefit from targeted therapies. And rapid advances in OMICS technologies and IT-based armamentarium, which identified individual differences, lead to produce diagnostic and screening biomarkers, targets and drugs of the principally new generations [7]. And, in addition, patient care will thus be revolutionized through the use of novel molecular predisposition, screening, diagnostic, prognostic, and monitoring markers. Although numerous challenges will need to be met to make PPM a reality, with time, this approach will replace the traditional trial-and-error practice of canonical medicine.
Figure 5A. Promise of OMICS to Personalized &
Precision Medicine
The ability to study biological phenomena at omics
levels in turn is expected to lead to significant advances
in PPM, which capitalizes on these conceptual and
technological advancements and stands on two main
pillars: data generation and data modeling.
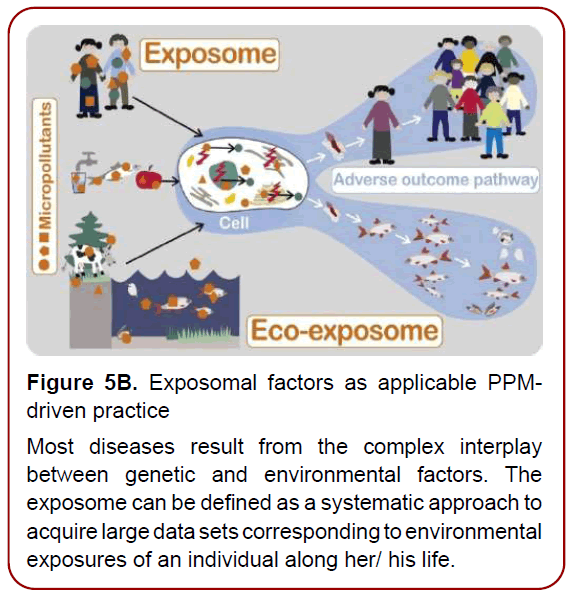
Figure 5B. Exposomal factors as applicable PPMdriven
practice
Most diseases result from the complex interplay
between genetic and environmental factors. The
exposome can be defined as a systematic approach to
acquire large data sets corresponding to environmental
exposures of an individual along her/ his life.
To determine molecular subtypes, ASD patients are first classified by applying clustering methods to different types of omics data, then these results are integrated with clinical data to characterize distinct disease subtypes. An example of this molecular-data-first approach is a spectrum of social communication disorders marked by tremendous etiological and phenotypic heterogeneity. In the case of ASD, OMICS-related data such as exome sequences and gene and protein expression data are combined with clinical data such as psychometric testing and imaging to enable subtype identification. In this sense, novel ASD subtypes have been proposed, such as CHD8, using this molecular subtyping approach.
Stratifying the sample by cluster analyses revealed quantitative differences in gene expression that appear to correlate with severity of ASD phenotype as well as gene expression profiles for each subtype that associate a “Biological phenotype” (i.e., gene expression profile) to the respective functional/behavioral phenotype. The biological phenotypes reveal differences in some of the biological functions affecting individuals with ASD, such as circadian rhythm dysregulation in the severe (L) phenotype, suggesting possible therapeutic interventions specific to this subgroup. On the other hand, overlapping genes among the phenotypes indicate dysregulation of genes controlling both neurological and metabolic functions that may lie at the core of ASD [8]. Broader use of molecular subtyping in complex disease research is impeded by data heterogeneity, diversity of standards, and ineffective analysis tools. The future of molecular subtyping for ASD and other complex diseases calls for an integrated resource to identify disease mechanisms, classify new patients and persons-at-risk, and inform effective preventive, prophylactic and therapeutic treatment options. This in turn will empower and accelerate PPM and PPM-driven healthcare services. Molecular subtypes need to be connected to rich, but easily attainable, clinical and phenotypic data so that molecular subtyping can be used to create personalized treatments for patients. The development of such resources to assist the molecular subtyping of ASD has the potential to accelerate both the classification of new patients and the development of treatment regimens tailored to the specific presentation of a given subtype. As a result, these resources will empower and accelerate PPM and will ultimately serve families in more proactive and comprehensive ways.
Development of high-throughput technologies has enabled scientists and clinicians to examine genomes, transcriptomes, proteomes, metabolomes, and other OMICS-related information in unprecedented detail. The combined ‘OMICS’ information leads to a global profiling of health and disease, and provides new approaches for personalized health monitoring and preventive treatment. To implement PPM resources into clinical practice, there is a strong need to develop a principally new strategy based on biomarkers, operating in the field of PPM and PPM-related subfields via sets Diagnostic, Predictive, Prognostic, On-Treatment and related ones (Figures6 a and b) [9]. Developing disease-specific biomarkers for identifying the stage of disease at the time of first diagnosis and for quantifying disease progression thereafter is an important requirement (Figure 7). Research in identifying and verifying disease-specific biomarkers would be extremely useful to manage the complexity of disease, as well as in enabling earlier detection of disease and hence making the advances in clinical practice and initiation of drug therapy.
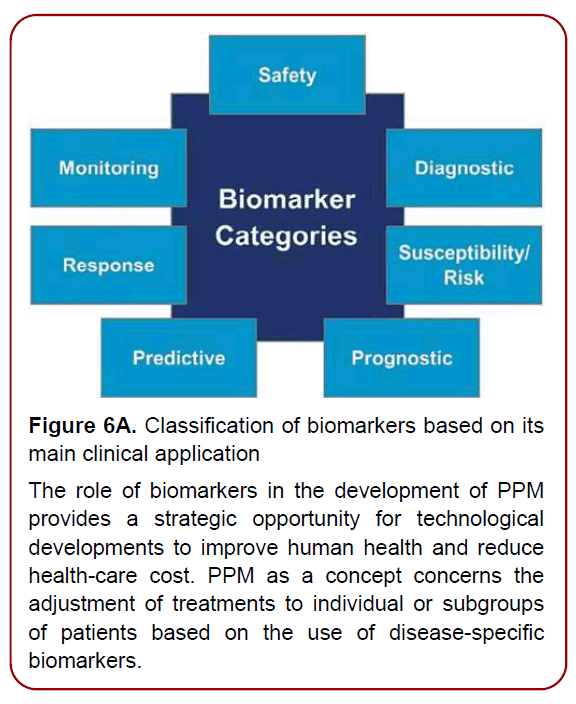
Figure 6A. Classification of biomarkers based on its
main clinical application
The role of biomarkers in the development of PPM
provides a strategic opportunity for technological
developments to improve human health and reduce
health-care cost. PPM as a concept concerns the
adjustment of treatments to individual or subgroups
of patients based on the use of disease-specific
biomarkers.
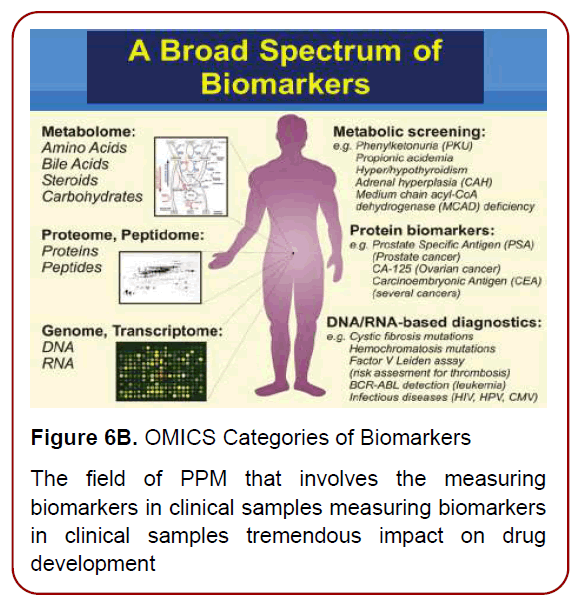
Figure 6B. OMICS Categories of Biomarkers
The field of PPM that involves the measuring
biomarkers in clinical samples measuring biomarkers
in clinical samples tremendous impact on drug
development
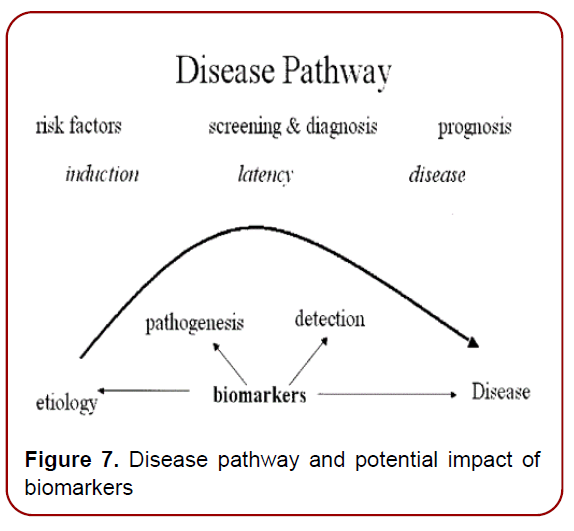
Figure 7. Disease pathway and potential impact of biomarkers
The directions of pathogenetic therapy within the framework of PMM
PPM can specifically target diseases depending on a person’s genetic makeup or environmental factors. It can predict how individuals will react to certain treatments, thus avoiding the ‘try it and see’ method used, in some cases, when prescribing mass-market drugs.
A biomarker-driven approach to developing targeted therapies (Figures 8a-8c) has the potential to increase success in the drug development process and ultimately improve patient outcomes.
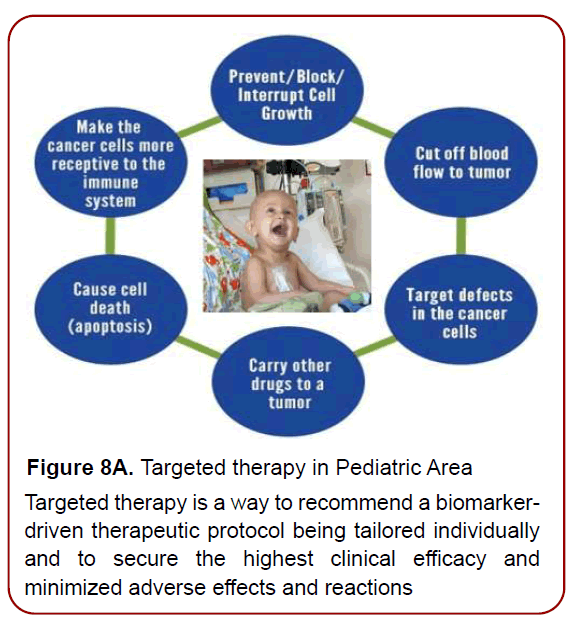
Figure 8A. Targeted therapy in Pediatric Area
Targeted therapy is a way to recommend a biomarkerdriven
therapeutic protocol being tailored individually
and to secure the highest clinical efficacy and
minimized adverse effects and reactions
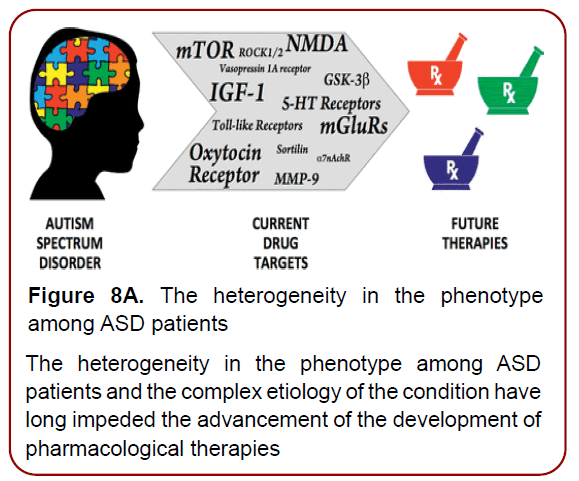
Figure 8A. The heterogeneity in the phenotype
among ASD patients
The heterogeneity in the phenotype among ASD
patients and the complex etiology of the condition have
long impeded the advancement of the development of
pharmacological therapies
Targeted treatments (targeted therapy)
It has been developed for several disorders that have a known specific genetic cause leading to autism. Since there are significant molecular and neurobiological overlaps among disorders, targeted treatments developed for a specific disorder may be helpful in ASD of unknown etiology. In this context, PPM-driven therapies will require the co-development of diagnostic & therapeutic tools to identify the optimal treatment for individual patients and/or persons-at-risk [10]. Most drug development now is aimed against a particular biomarker-related molecular target, and many drugs will primarily benefit patients and/or personsat- risk with a specific molecular abnormality. These may include genetic mutations, dysregulated biomarkers, or even tissue-specific gene expression signatures, or even interactome-, network- and pathway-related abnormalities. And thus, a key point of special interest is becoming in clinical practice a biomarker test which would have to be ethically acceptable.
The success of PPM-driven approaches has rested on several pillars, each of which is updated and enhanced with new technologies:
1. genotypic and OMICS-driven studies,
2. patient stratification, and
3. iterative research process.
Although PPM and related modern resources have become a genuine reality for practitioners and for a wide swath of patients as well, there remains much promise of PPM in many areas of disease (with full clinical manifestations) and/or in pre-illness conditions (at subclinical stage), and possibly none more so than in neurodevelopmental disorders (NDDs). The challenges in this therapeutic area are intrinsically linked to the complex biology of NDDs and the vulnerability of the patient populations. And despite the shared features defining the ASD phenotype, a more PPM-driven approach is currently recommended for individuals with autism spectrum disorder (ASD) (Figure 9). ASD is a neurodevelopmental disorder that manifests during the early development of child’s brain as persistent impairments in social interaction and incessant, repetitive behaviors. Moreover, ASD is a kind of disorder with a high prevalence but a very limited armamentarium of targeted drug-driven interventions. This paradox may be partly explained by the heterogeneity of the affected population and limited understanding of etiology in the majority of cases. At the same time, the number of drug-driven treatments approved for ASD is quite low and exclusively aimed at management of the associated symptom domains. No disease-modifying therapies targeting the core symptom domains are currently available. Although a majority of recent and ongoing clinical trials in ASD include PPM-driven approaches, the lack of established relationships between biomarkers, biomarker-driven and supported targets, and clinical outcomes persists. So, following the principles of PPM, patient stratification based on genetics, OMICS-supported datasets, clinical features, or biomarker profiles is considered to become a promising approach that may link novel (targeted and/or “smart”) treatments to individual patient cohorts in a more precision-based approach.
Figure 9. Autistic Spectrum Disorder (ASD) is a
developmental disorder which mainly affects a child’s
development of communication and socialization
skills
Children with ASD also may exhibit very challenging
behaviour and anxiety
So, being summarized, ASD as a kind of neuronal, metabolic and heterogeneous disorders caused by an interaction between genetic vulnerability and environmental factors, is characterized by a panel of highly informative biomarkers, including oxidative stress-, decreased methylation capacity-, limited trans-sulfuration production of cysteine and GSH, mitochondrial dysfunction, intestinal dysbiosis, increased toxic metal burden, cerebral hypo-perfusion, and complex immune dysregulation [11]. No comments, but therapeutic interventions being based on biomarker-driven research and applications, are most effective if started pre-early in life (at subclinical stage), yet diagnosis often remains delayed, partly because the diagnosis of ASD is based on identifying abnormal behaviors that may not emerge until the disorder is well established. Biomarkers that identify children at risk during the pre-symptomatic period, assist with pre-illness diagnosis, confirm behavioral observations, stratify patients into subgroups, and predict therapeutic response would be a great advance.
However, most biomarkers have not undergone validation studies and most studies do not investigate biomarkers with clinically relevant comparison groups. Although the field of biomarker research in autism spectrum disorder is promising, it appears that it is currently in the early stages of development.
Biomarker-related research is already moving in this direction, as evidenced by studies that have developed biomarkers such as maternal autoantibody-related autism biomarkers, which can predict ASD prenatally, and genetic biomarkers, which may link specific syndromes to ASD. Studies of metabolic biomarkers may provide insight into treatment, while neuroimaging biomarkers may provide biologically relevant indicators of abnormal brain development. Meanwhile, regardless to the promising prospective, ASD is currently diagnosed using, unfortunately, only behavioral criteria. The formal diagnosis of ASD requires clinical expertise and performance of lengthy “gold standard” tools, which can lead to a suboptimal rate of diagnosis. The significant delays in age of diagnosis and discrepancies in availability of diagnostic, predictive and prognostic tools, intervention services, and in the level of professional expertise are a significant challenge. And the identification of ASD is thus pre-required for provision of appropriate intervention and treatment.
Early intervention may lessen the severity of the autistic manifestations in children with ASD. Studies have shown that 3-25% of children with early and pre-early (subclinical) diagnoses reach normal levels of social, adaptive, and cognitive. Interestingly, a randomized clinical trial showed remarkable improvements in the core features and adaptive behaviors of ASD after two years of a behavioral intervention performed at thirty months of age or earlier. Unfortunately, many ASD patients are diagnosed at an older age. A recent meta-analysis showed that the mean age at diagnosis was 60.48 months after reviewing thirtyfive studies performed on fifty-five cohorts collectively comprising almost sixty-seven thousand ASD patients from thirty-five countries. Therefore, the goal in current practice is to identify ASD as early as possible to promote pre-early intervention and better outcomes (Figure 10) [12]
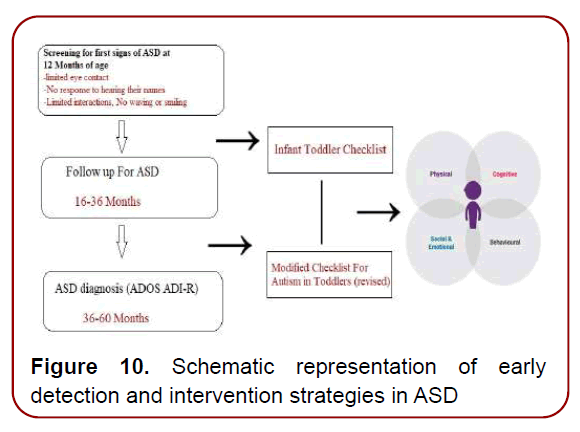
Figure 10. Schematic representation of early detection and intervention strategies in ASD
Challenges for ASD diagnoses and interventions
Although ASD may not be fully manifested before school age or a later stage, the symptoms may begin to appear between six and eighteen months of age. Heterogeneity of clinical presentation and etiology of ASD have hindered the discovery of risk factors and pathophysiological underpinnings of this umbrella disorder. And the individualized precise genetic risk factors (Figure 11) are the contribution of our genes to play in the chance we have of developing certain illnesses or diseases, including ASD [13]. However, a number of individual high-risk genes have been identified in studies where specific de novo mutations were strongly associated with NDDs characterized by a high prevalence of autism. And only now we are about to understand the architecture of the human gene networks in ASD and ultimately a new hope for patients affected by these genetic variants [14]. Meanwhile, identifying the brain temporo-spatial regions where the risk genes are expressed in ASD patients may help to improve the therapeutic strategies. ASD-Risk used an optimal feature selection algorithm called inheritable bi-objective combinatorial genetic algorithm to identify the brain temporo-spatial regions for prediction of the risk genes of ASD (Figure 12). The formal diagnosis of ASD requires clinical proficiency and meeting an extensive set of “gold standard” criteria, which can substantially reduce early identification of ASD. Difficulties in accessing diagnostic tools, intervention facilities, and training experts and specialists, are global challenges in the diagnosis of ASD and can increase the age of diagnosis. The final diagnosis of ASD is required for proper therapeutic and intervention plans as well.
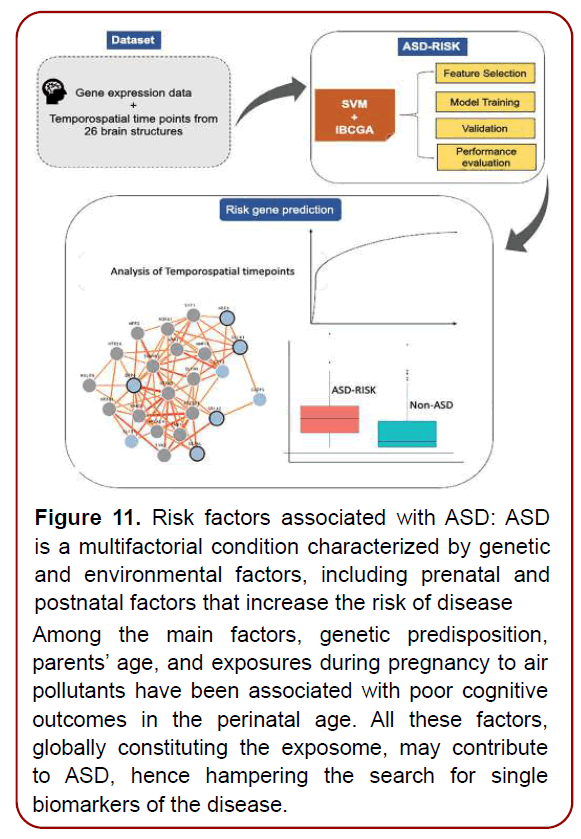
Figure 11. Risk factors associated with ASD: ASD is a multifactorial condition characterized by genetic and environmental factors, including prenatal and postnatal factors that increase the risk of disease Among the main factors, genetic predisposition, parents’ age, and exposures during pregnancy to air pollutants have been associated with poor cognitive outcomes in the perinatal age. All these factors, globally constituting the exposome, may contribute to ASD, hence hampering the search for single biomarkers of the disease.
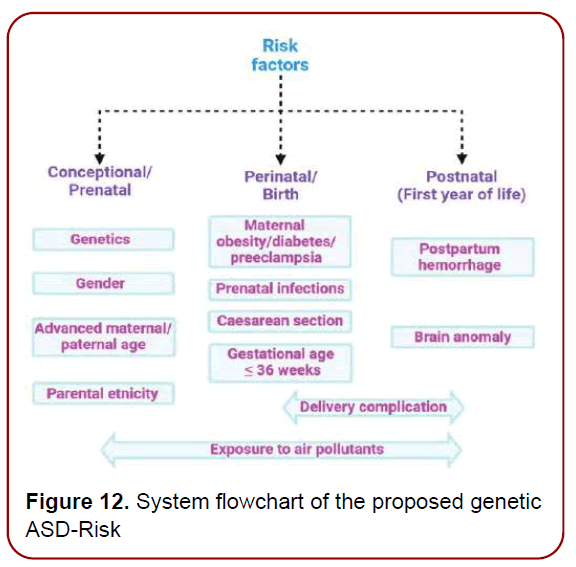
Figure 12. System flowchart of the proposed genetic ASD-Risk
In this context, genomics is considered to be a set of the unique biomarkers and thus the molecular tools to probe genome for its quality and now even be tested! Genetics & Genomics can thus be used to assess the etiological roles of biomarkers in disease and to prioritize drug targets, including designing their evaluation in clinical trials. So, genomics is thus a source of Predictive and Prognostic biomarkers (Figure 13) on one hand, and constructing genome-driven medicines to improve genome landscape, on the other one. The latter sounds much crucial for the future of ASD-related disorders.
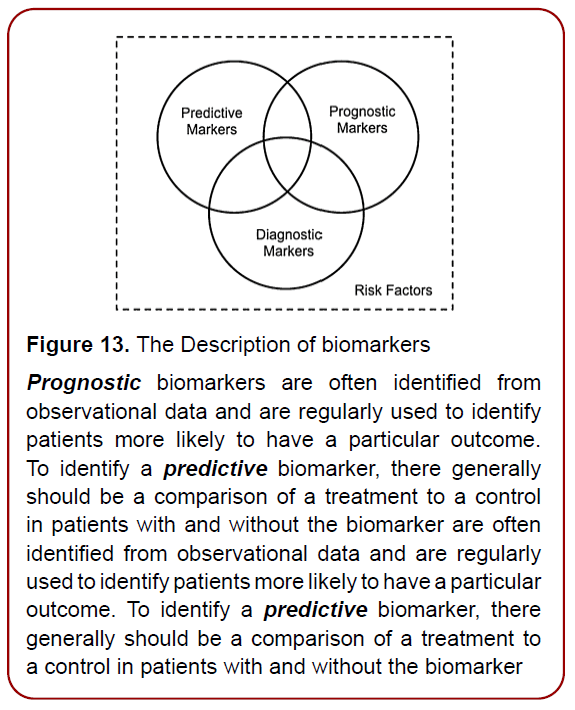
Figure 13. The Description of biomarkers
Prognostic biomarkers are often identified from
observational data and are regularly used to identify
patients more likely to have a particular outcome.
To identify a predictive biomarker, there generally
should be a comparison of a treatment to a control
in patients with and without the biomarker are often
identified from observational data and are regularly
used to identify patients more likely to have a particular
outcome. To identify a predictive biomarker, there
generally should be a comparison of a treatment to
a control in patients with and without the biomarker
As you might see, genetic studies are illuminating the molecular pathophysiology of ASD, and new tools such as induced pluripotent stem cells offer novel possibilities for drug screening and disease diagnostics. And thus, the large-scale collaborations between academia and bioindustry are starting to address some of the key barriers to developing drugs for ASD [15].
Having access to the deepest genomic information (Figures14 a and b), via unique genomic technologies and genomic testing will become increasingly important as physicians are progressively receptive to incorporating genomics into clinical (including Neurology-related) practice.
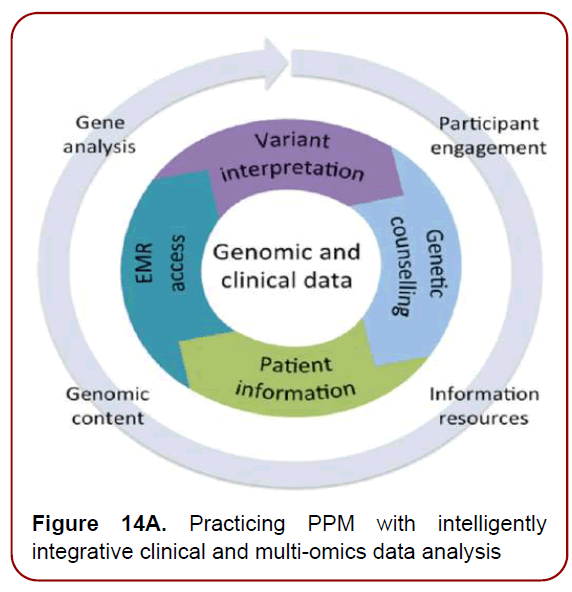
Figure 14A. Practicing PPM with intelligently integrative clinical and multi-omics data analysis
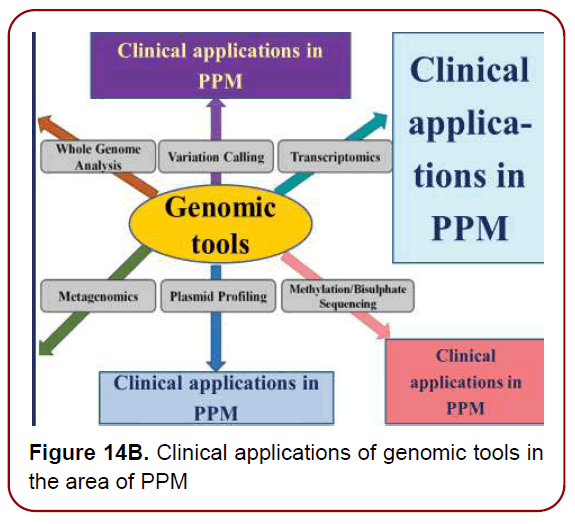
Figure 14B. Clinical applications of genomic tools in the area of PPM
In this context, genetic testing can provide information about person’s genes, their products and chromosomes. In brief, genetic tests and genome profiling can help to:
• Diagnose, prognosticate or predict disorder
• Identify gene changes that are responsible for an already diagnosed disease
• Guide doctors in deciding on the best medicine or treatment to use for certain individuals
• Identify gene markers and loci to be used for biomarkerdriven targeted drug discovery
• Screen newborn babies for certain treatable conditions and to develop the targeted medicines
For instance, identifying genetic determinants of common disease through genome-wide association studies (GWAS) & phenome-wide association studies (PheWAS) (Figure. 15)
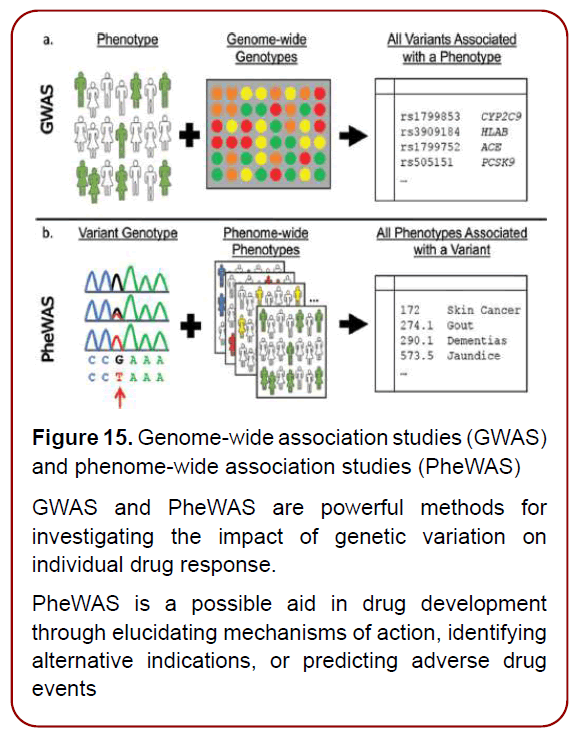
Figure 15. Genome-wide association studies (GWAS)
and phenome-wide association studies (PheWAS)
GWAS and PheWAS are powerful methods for
investigating the impact of genetic variation on
individual drug response.
PheWAS is a possible aid in drug development
through elucidating mechanisms of action, identifying
alternative indications, or predicting adverse drug
events
provided valuable knowledge about the genetic architecture of disease and potential pointers to underlying causative mechanisms and engendered the promise of discovering better candidate targets for common, complex diseases (such as ASD, diabetes or atherosclerosis), cancer and autoimmune conditions. GWAS-PheWAS tandem has provided powerful methods for investigating the impact of genetic variation on individual drug response and have added extensive knowledge to the understanding of drug targets and effects [16]. In this connection, next-generation sequencing (NGS) technologies (Figure. 16)
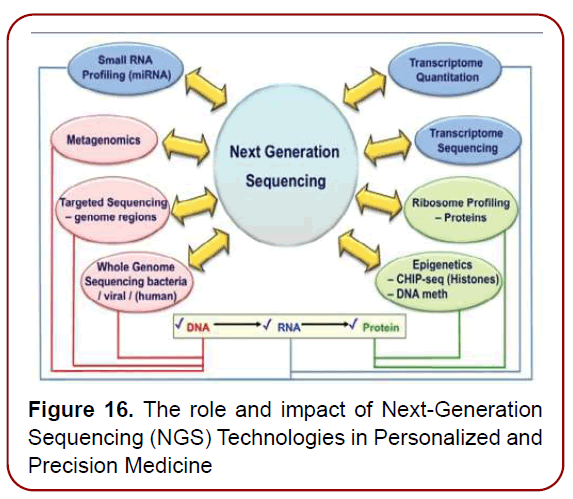
Figure 16. The role and impact of Next-Generation Sequencing (NGS) Technologies in Personalized and Precision Medicine
Next-generation sequencing (NGS): technology among the genomic profiling technologies allows for the rapid and accurate sequencing of many genes at once. NGS holds great promise for unravelling the mysteries of biosystems, with the advent of DNA/RNA sequencing methods of the next-step generation having greatly accelerated biomedical research, gaining broad applicability in disease diagnosis and therapeutics in recent years and future to come [17]. This technology is becoming more common in clinical practice, though the clinical benefit of incorporating it into PPM-driven strategies remains under significant debate.
Diagnostic testing: It is used to identify or rule out a specific genetic or chromosomal condition. The results of a diagnostic test can influence a person’s choices about health care and the management of the disorder.
Presymptomatic and predictive testing: can be helpful to people who have a family member with a genetic disorder, but who have no features of the disorder themselves at the time of testing. The latter can identify mutations that increase a person’s risk of developing disorders with a genetic basis, such as MS, ASD, etc.
Carrier testing: is offered to individuals who have a hidden (latent) viral gene pre-integrated into the human genome or to people in certain ethnic groups with an increased risk of specific genetic conditions.
Newborn testing: is to identify genetic disorders (including ASD) that can be treated early in life and other disorders including monogenic and orphan diseases.
Pre-conception testing: (Figure 17) is an important tool for couples whose genetic make-up might have negative consequences on their children’s health.
Figure 17. Pre-conception testing
Prenatal testing: is to detect changes in a fetus’s genes or chromosomes before birth to secure the national biosafety.
Preimplantation testing: (also called preimplantation genetic diagnosis (PIGD) can reduce the risk of having a child with a particular genetic or chromosomal disorder.
Pharmacogenomics-related testing (Figure 18 a and b). is aimed at tailoring drug therapy at a dosage that is most appropriate for an individual patient, with the potential benefits of increasing the clinical efficacy and individualized safety
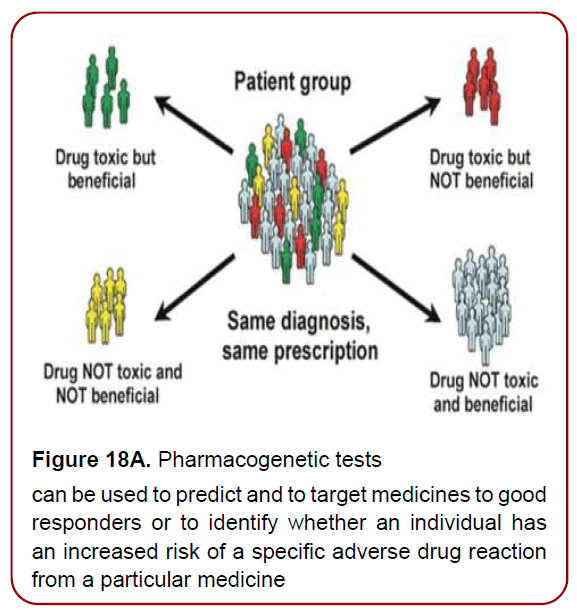
Figure 18A. Pharmacogenetic tests
can be used to predict and to target medicines to good
responders or to identify whether an individual has
an increased risk of a specific adverse drug reaction
from a particular medicine
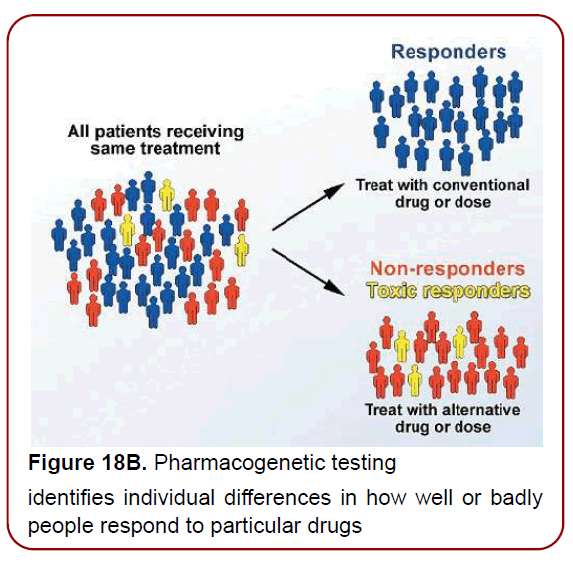
Figure 18B. Pharmacogenetic testing
identifies individual differences in how well or badly
people respond to particular drugs
Nutrigenomics testing is a specialty of the interaction between genes and nutrients whilst prompting to get the Individualized Diet set up. It suggests that an individual’s diet should be matched to their genetic makeup (Figure 19)
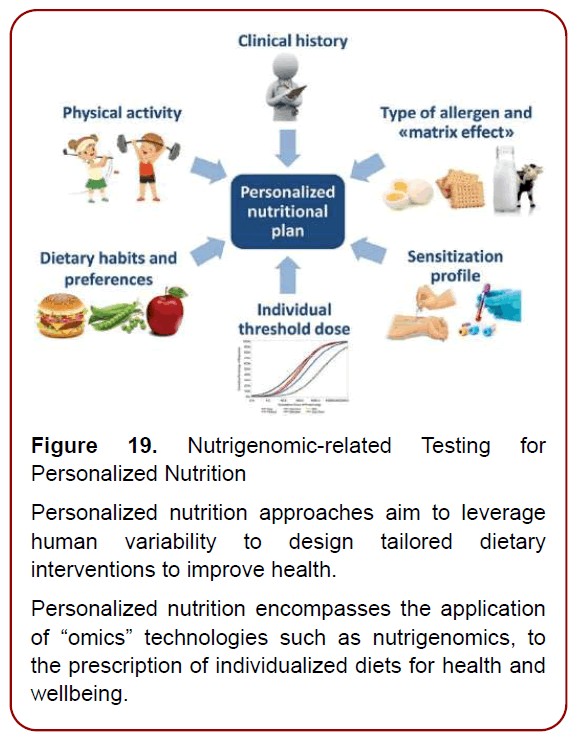
Figure 19. Nutrigenomic-related Testing for
Personalized Nutrition
Personalized nutrition approaches aim to leverage
human variability to design tailored dietary
interventions to improve health.
Personalized nutrition encompasses the application
of “omics” technologies such as nutrigenomics, to
the prescription of individualized diets for health and
wellbeing.
The landscape of genetic testing has changed considerably with the emergence of direct-to-consumer (DTC) genetic testing (Figure 20) to secure individualized genetic risk prediction tools for a wide array of common diseases (including ASD patients and persons-at-risk) and thus the national health stability [18]

Figure 20. Direct-to-consumer (DTC) genetic testing set
The day when children (ASD-related persons-at-risk, in particular) visit their pediatricians, get genome scans, and leave with personalized plans for their health care over the subsequent 20 years remains distant. However, it is important to recognize that genotyping and low-cost sequencing have created an inflection point in the pace of genomic discovery relevant to clinical care. Moreover, the genomic diagnostic evaluation of undiagnosed ASD, and reproductive genetics/newborn screening exemplify the need for the practicing pediatrician to have competency in genomics. Pediatricians have the unique and exhilarating responsibility to help ensure that young ASD patients and/ or persons-at-risk derive maximal benefit from genomics which, in turn, will provide pediatricians new and often unexpected insights into the biological basis of health and disease and will afford new health care options requiring informed and sometimes challenging choices of physicians and patients.
Despite these efforts, there is no consensus on the best diagnostic, prognostic, or predictive (for available interventions) biomarkers yet. The difficulties in identification and implementation of the ASD biomarkers are reflected in the lack of ASD-related biomarker-driven targeted therapeutics.
A majority of the high-risk ASD genes are expressed during brain development with roles in transcriptional regulation, chromatin remodeling, synaptic function, and neuronal communication. Noteworthy is the fact that the number of implicated genes far exceeds the number of clinically defined phenotypes. Thus, multiple genes are likely to converge on shared pathways, resulting in a similar ASD phenotype. And ‘‘mapping’’ high-risk genes to biological networks, interactomes and pathways can be conducted in various models and further validated.
Meanwhile, considerable evidence is emerging that ASD is most often triggered by a range of different genetic variants that interact with environmental (exposomal) factors (Figure. 21). To date, many different approaches have been used to investigate the genetics of ASDs, from gene or chromosome-focused linkage analyses and familybased association tests to the more recent genome-wide linkage and association analyses, which have interrogated hundreds of thousands of SNPs in thousands of probands and families. These studies have together resulted in the Up to 80% of genetic variations that contribute to ASD found to date are neither extremely rare nor classified as pathogenic. Rather, they are less common single nucleotide polymorphisms (SNPs), found in 1-15% or more of the population, that by themselves are not disease-causing. These genomic variants contribute to ASD by interacting with each other, along with nutritional and environmental factors. And PPM, as it is understood, as an upgraded Model of Healthcare Services of the next step generation and being a Science and ART, illustrates application of sets of the different tools at the population, community-related and individual levels, whilst exerting reliable control over morbidity, mortality and disabling rates as well as significantly optimizing the cost and efficacy of treatment for those who had fallen ill –patients, and for persons-at-risk [19].
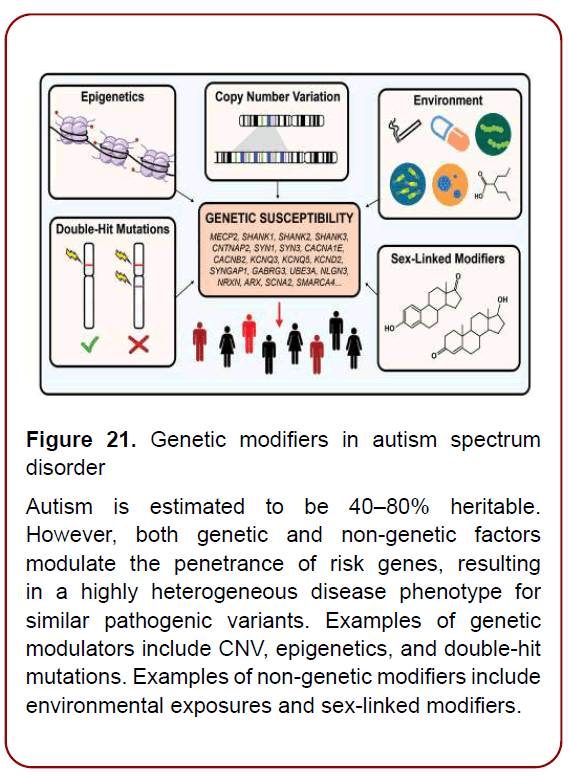
Figure 21. Genetic modifiers in autism spectrum
disorder
Autism is estimated to be 40–80% heritable.
However, both genetic and non-genetic factors
modulate the penetrance of risk genes, resulting
in a highly heterogeneous disease phenotype for
similar pathogenic variants. Examples of genetic
modulators include CNV, epigenetics, and double-hit
mutations. Examples of non-genetic modifiers include
environmental exposures and sex-linked modifiers.
Right now, massive genetic studies are currently underway producing data to implicate additional genes. Meanwhile, numerous biomarker studies using a wide range of modalities have attempted to identify - and some to fully validate – a variety of biomarker candidates in ASD. Standardization of the biomarker methodologies, including parsing heterogeneity and well-controlled crosssectional and longitudinal studies, will be required to evaluate relationships between biomarker changes and clinical improvements. In-depth genotype-phenotype analyses, as well as quantitative in vitro and in vivo ASD models, will be key for discovery of new biomarker-driven targeted therapeutics or repurposing of the existing ones. Of particular interest in this sense are in vitro models developed by using patient-derived pluripotent stem cells (PPSCs) to generate neuronal cultures or organoids. This approach affords a unique opportunity to study biological processes in the context of neuronal function and in the genetic background of affected individuals. These models can be used for identification of potential therapeutics in two major ways: (i) phenotypic screening or (ii) testing of drug candidates targeting specific molecular entities [20]. The PPM-driven approach may improve the biomarker-supported diagnosis and long-term prognosis in ASD. Earlier (including subclinical) detection of ASD enables earlier or critical time point intervention, which, in turn, is associated with improved long-term prognosis. A key improvement may be to establish a correlation between biomarkers readily assessable in infants; for example, pupil dilation fluctuations, and a gold-standard diagnosis of ASD (by validated molecular tools and clinical instruments) later in life.
This work was funded through the National Plan for Science Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia (award number:08-MED 510-02).
The authors wish to acknowledge the National Plan for Science Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia (award number:08-MED 510-02).
AE: Suggested the topic and drafted the manuscript; LA: Carefully revised and has approved the submitted version
This work was approved by the ethics committee of King Khalid Hospital, King Saud University (Approval number: 11/2890/IRB).
Not applicable
The authors declare no conflict of interest. The sponsors had no role in the design, execution, interpretation, or writing of the study.